| 2003 | Dr. sc. nat., Institute of Biochemistry, ETH Zurich, CH |
| 2004-2007 | Postdoc MRC Laboratory of Molecular Biology, Cambridge, UK |
| 2007-present | Group Leader, ZMBH |
|
|
Marius Lemberg's Laboratory
|
|
|
|
| Mechanism and function of intramembrane proteases in physiology and molecular aging
Intramembrane proteolysis has over the last few years become increasingly recognised as an important signalling mechanism with functions ranging from transcription control to regulated growth factor secretion. They have also been implicated in important diseases such as Alzheimer's and infection by pathogens including hepatitis C virus, and just recently a mitochondrial intramembrane protease has been implicated in apoptosis. We are interested to elucidate the function and roles of these proteases in physiologigical processes and in events of molecular aging. Intramembrane proteases comprise three mechanistic classes. The first identified example, the site-2 protease, is a metalloprotease. The second family comprising seven members in humans is the group of aspartyl proteases including gamma-secretase, signal peptide peptidase (SPP) and SPP-like protease. The third family comprises the rhomboid serine proteases with five active members in humans. Intramembrane proteolysis in the eukaryotic secretory pathway commonly results in the selective release of bioactive signalling molecules. Despite a few well-studied examples such as gamma-secretase, very little is known about the function of most of the 13 intramembrane proteases known in humans and mice. Although the recent progress in the biochemical analysis of solubilized intramembrane proteases, and most recently a number of high-resolution structures of rhomboid intramembrane proteases, has led to first real insights, key mechanistic questions remain. My lab is interested in understanding how these mysterious proteases work on the molecular level and how they act on cellular membrane homeostasis and cell death. By combining various approaches ranging from biochemistry, cell biology to bioinformatics we aim to identify their substrates and analyze their function. My group is supported by a grant from Landesstiftung Baden-Wuerttemberg. Lemberg MK, Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 17: 1634-1646. (2007) Lemberg MK, Menendez J, Misik A, Garcia M, Koth CM, Freeman M. Mechanism of intramembrane proteolysis investigated with purified rhomboid proteases. EMBO J. 24: 464-472. (2005) Lemberg MK, Martoglio B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10:735-744. (2002) Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296: 2215-2218. (2002) Lemberg MK, Bland FA, Weihofen A, Braud VM, Martoglio B. Intramembrane proteolysis of signal peptides: an essential step in the generation of HLA-E epitopes. J. Immunol. 167: 6441-6446. (2001) Reviews Lemberg, MK, Freeman M. Cutting proteins within lipid bilayers: rhomboid structure and mechanism. Mol. Cell. 28: 930-940. (2007) Lemberg MK, Martoglio B. On the mechanism of SPP-catalysed intramembrane proteolysis; conformational control of peptide bond hydrolysis in the plane of the membrane. FEBS Lett. 564: 213-218. (2004) |
|
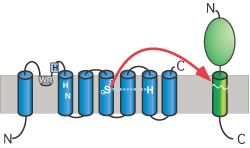
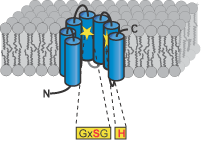
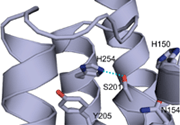
Figures: Top panel, topology model for secretase-type rhomboids with the position of the functional important and highly conserved residues highlighted. Typical for all intramembrane-cleaving pro-teases, rhomboids have their active site residues within transmembrane regions. This is consistent with the observed cleavage of substrates in their transmembrane portion; mul-tiple transmembrane regions assemble a proteolytic domain within the plane of the membrane (cartoon, middle panel). The recently solved crystal structure of the E. coli rhomboid GlpG reveales that the serine histidine catalytic dyad is located in an aqueous pocket 10 Å below the membrane surface (ribbon diagram using coordinates from Wang, Zhang and Ha, Nature 2006); see Lemberg and Freeman (Mol Cell, 2007) for review.