|
Welcome to the Bukau Lab!
Biogenesis & quality control of proteins
The ensemble of molecular chaperones are central components of the cellular machinery that establishes and maintains protein homeostasis. Chaperones assist native folding of newly synthesized proteins and repair and eliminate misfolded and aggregated proteins, and therefore have fundamental impact on cell physiology, aging and disease. The goal of our research is to understand the intricate functional network of chaperones and its interplay with proteases in protein biogenesis and quality control, and the molecular working principles of chaperone machines. Furthermore, we want to elucidate causes and consequences of protein aggregation related to disease, in particular neurodegeneration and cancer. As models we are using E. coli, S. cerevisiae, C. elegans and human cells, and we are employing multi-disciplinary approaches ranging from genetics and molecular biology to biochemistry and biophysics. Currently we have three main research themes:
1. Mechanisms of folding and assembly of newly synthesized proteins.
Cells from bacteria to humans have evolved a multilayered machinery that engages translating ribosomes to promote folding and assembly of newly synthesized proteins. Using ribosome profiling, genetics and protein biochemistry, we want to understand how this machinery guides nascent polypeptides to the native state, and how assembly of oligomeric protein complexes is achieved in pro- and eukaryotes.
2. Mechanisms of protein quality control.
Disrupting proteostasis of living cells activates protective quality control systems, which refold or degrade misfolded proteins or sequester potentially cytotoxic misfolded proteins into aggregates, deposited at specific intracellular sites. We want to understand the cellular processes leading to targeted deposition of aggregating proteins inside cells. We are also dissecting the mechanisms by which the Hsp70 chaperone network and the AAA+ disaggregase Hsp104 solubilize and refold aggregated proteins, including disease-associated amyloid fibrils.
3. Propagation of protein misfolding in neurodegenerative diseases (Project group Carmen Nussbaum-Krammer).
Neurodegenerative diseases exhibit a complex pathology involving non-cell autonomous effects and progressive spreading of protein misfolding. Using the metazoan model system C . elegans we want to understand how local protein misfolding is affecting neighboring cells and tissues and how proteostasis is orchestrated at the organismal level.
Selected Publications
Original Papers
Bertolini, M., Fenzl, K., Kats, I., Wruck, F., Tippmann, F. Schmitt, J., Auburger, J.J., Tans, S., Bukau, B., Kramer, G. Interactions between nascent proteins translated by adjacent ribosomes drive homomer assembly. (2021) Science 371: 57-64, https://doi.org/10.1126/science.abc7151, see Abstract, Reprint, Full text.
Wentink, A.S. et al. Molecular dissection of amyloid disaggregation by the human Hsp70 chaperone. (2020) Nature, https://doi.org/10.1038/s41586-020-2904-6.
PNAS Journal Club: "Insights into heat shock protein machinery could point to interventions for neurodegenerative disease" by Amy McDermott, 20 November 2020 (read more).
Faust, O. et al. Hsp40 proteins use class-specific regulation to drive Hsp70 functional diversity. (2020) Nature, https://doi.org/10.1038/s41586-020-2906-4.
PNAS Journal Club: "Insights into heat shock protein machinery could point to interventions for neurodegenerative disease" by Amy McDermott, 20 November 2020 (read more).
Avellaneda, M.J. et al.Processive extrusion of polypeptide loops by a Hsp100 disaggregase. (2020) Nature, doi: 10.3390/ijms20246220. (Abstract), Erratum.
Ho, C.-T. et al. Cellular sequestrases maintain basal Hsp70 capacity ensuring balanced proteostasis. (2019) Nature Commun. 10:4851. doi: 10.1038/s41467-019-12868-1. (Abstract).
Shiber, A. et al. Co-translational assembly of protein complexes in eukaryotes revealed by ribosome profiling. (2018) Nature, doi: 10.1038/s41586-018-0462-y. (Abstract)).
Döring, K. et al. Profiling Ssb-nascent chain interactions reveals principles of Hsp70-assisted folding. (2017) Cell 170:298-311.e20. doi: 10.1016/j.cell.2017.06.038. (Abstract).
Ungelenk, S. et al. Small heat shock proteins sequester misfolding proteins in near-native conformation for cellular protection and efficient refolding. (2016) Nat. Commun. 7, 13673 (Abstract).
Schibich, D. et al. Global profiling of SRP interaction with nascent polypeptides. (2016) Nature 536, 219-223, doi: 10.1038/nature19070 (Abstract).
Shieh, Y.-W. et al. Operon structure and cotranslational subunit association direct protein assembly in bacteria. (2015) Science 350, 678 - 680 (Abstract).
Gao, X. et al. Human Hsp70 Disaggregase Reverses Parkinson's-Linked x-Synuclein Amyloid Fibrils. (2015) Mol. Cell. 59, 781-793. doi: 10.1016/j.molcel.2015.07.012 (Abstract).
Nillegoda et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. (2015) Nature 524, 247 - 251, doi:10.1038/nature14884 (read more).
Miller, S.B. et al. Compartment-specific aggregases direct distinct nuclear and cytoplasmic aggregate deposition. (2015) EMBO J. 34, 778-797. (Abstract)
Reviews
Ruger-Herreros, C. , Svoboda, L., Mogk, A., Bukau, B. Role of J-domain Proteins in Yeast Physiology and Protein Quality Control. (2024) J Mol Biol. doi: 10.1016/j.jmb.2024.168484. (Abstract).
Rosenzweig et al. The Hsp70 chaperone network. (2019) Nat. Rev. Mol. Cell Biol. doi: 10.1038/s41580-019-0133-3. (Abstract).
Wentink, A., Nussbaum-Krammer, C., Bukau, B. Modulation of Amyloid States by Molecular Chaperones. (2019) Cold Spring Harb Perspect Biol., pii: a033969. doi: 10.1101/cshperspect.a033969 (Abstract).
Mogk et al. Cellular Functions and Mechanisms of Action of Small Heat Shock Proteins. (2019) Annu. Rev. Microbiol. doi: 10.1146/annurev-micro-020518-115515 (Abstract).
Kramer et al. Mechanisms of Co-Translational Maturation of Newly Synthesized Proteins. (2018) Ann. Rev. Biochem., doi: 10.1146/annurev-biochem-013118-111717. (Abstract).
Nillegoda, N.B., Wentink, A.S., Bukau, B. Protein Disaggregation in Multicellular Organisms. (2018) Trends Biochem. Sci. pii: S0968-0004(18)30024-0. doi: 10.1016/j.tibs.2018.02.003. (Abstract).
Mogk et al. Cellular handling of protein aggregates by disaggregation machines. (2018) Mol. Cell 69:214-226. doi: 10.1016/j.molcel.2018.01.004. (Abstract).
|
|
 see the Bukau lab´s twitter account see the Bukau lab´s twitter account
 Bertolini, M., Fenzl, K. et al., Science (2021)
Image by Marzia Munafo
The ways to cotranslational protein complex assembly
Bertolini, M., Fenzl, K. et al., Science (2021)
Image by Marzia Munafo
The ways to cotranslational protein complex assembly
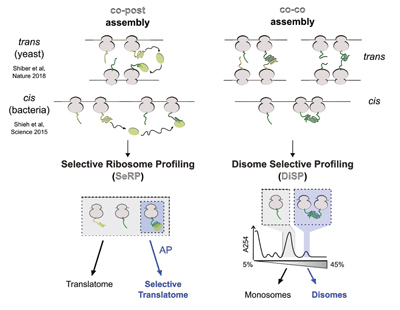 Bertolini, M., Fenzl, K. et al., Science, (2021)
Co-translational folding and assembly in yeast
Bertolini, M., Fenzl, K. et al., Science, (2021)
Co-translational folding and assembly in yeast
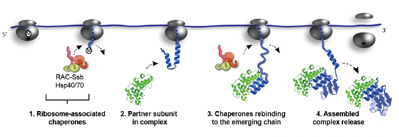
Döring, K., et al., Cell (2017)
Hsp70-mediated disassembly of amyloids
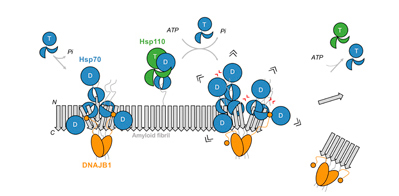 Wentink et al., Nature (2020) and Faust et al., Nature (2020), in press
Disassembly of amyloids by the human Hsp70 machinery
Wentink et al., Nature (2020) and Faust et al., Nature (2020), in press
Disassembly of amyloids by the human Hsp70 machinery
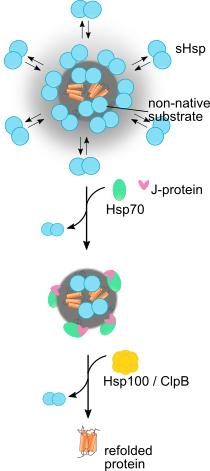
Mogk et al., Annu. Rev. Microbiol. (2019)
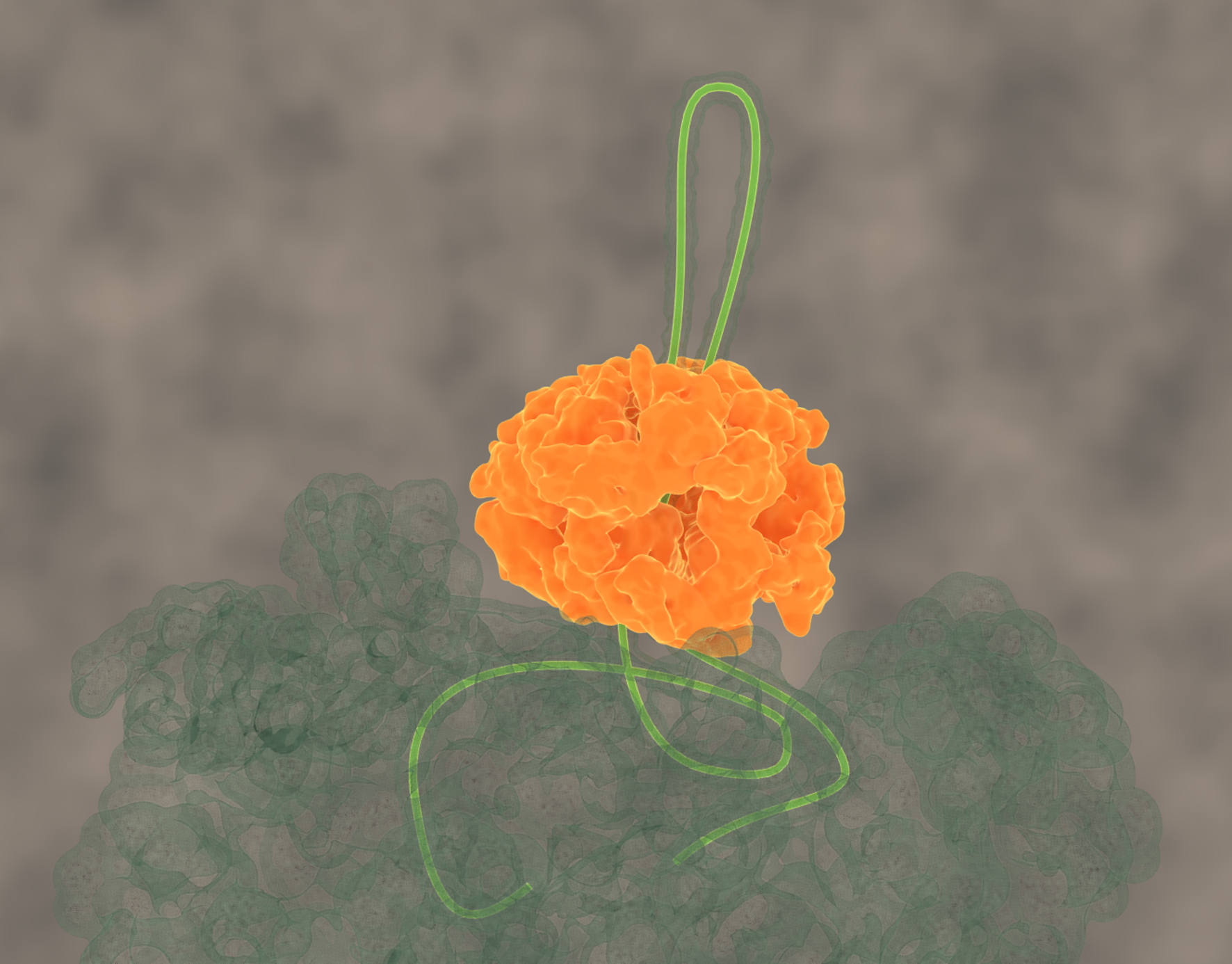
Protein disaggregation by the ClpB threading machine. (click for youtube movie)
|

