The MCB Curriculum
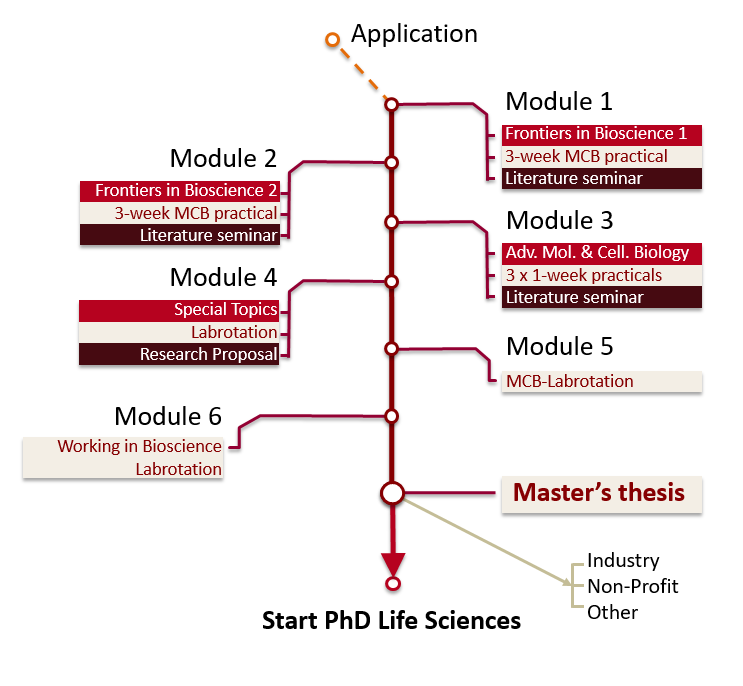
The programme starts at the beginning of October and consists of 4 semesters (2 years). However, with careful planning, it can also be completed in 18 months. In the first semester, you will attend two lecture courses called Frontiers in Bioscience 1 and 2 that are provided for all MSc students in biological sciences. At the same time you will participate in two advanced (3 week) practical courses including seminars. The second semester starts with Frontiers in Methodology, a module that concentrates on techniques; in addition to lectures, you choose three one-week practicals from a wide range of options. The fourth module offers the opportunity to learn about the research going on in the research groups associated to the MCB programme, and to practice writing a research proposal; at the same time you will undertake a 6-week project in a laboratory of your choice. Two further, 8-week laboratory practicals will enable you to widen your experience and choose the lab in which you would like to complete your 6-month master's thesis. Throughout the programme, you will participate in literature and research seminars in which you can learn to critically assess research papers and improve your language and presentation skills.
Click on the modules below to learn more about the contents.
Every module is worth 15 credits (ECTS), see this diagram for more details.