Contact us:
Zentrum für Molekulare Biologie der
Universität Heidelberg (ZMBH)
Im Neuenheimer Feld 345
69120 Heidelberg, Germany
Tel.: +49-6221-54 6813
Fax: +49-6221-54 5894
ms-service@zmbh.uni-heidelberg.de
|
PTM analyses We
perform the analyses of various
posttranslational modifications (PTMs), for
example phosphorylation1,
SUMOylation, gykosylation2,3, cystein
redox state4
etc. Please contact us for details and to
discuss your specific project. See also the
exemplary citations at the end of the page.
Phosphorylations
We can perform large scale/global
phosphorylation analysis from complex samples
(Figure 1), which always involves a
phosphopeptide enrichment step, to account for
the low number of phosphorylated peptides
compared to unphosphorylated ones (~1%).
Moreover, to further reduce sample complexity,
offline peptide fractionations (SCX, high-pH)
can be employed.
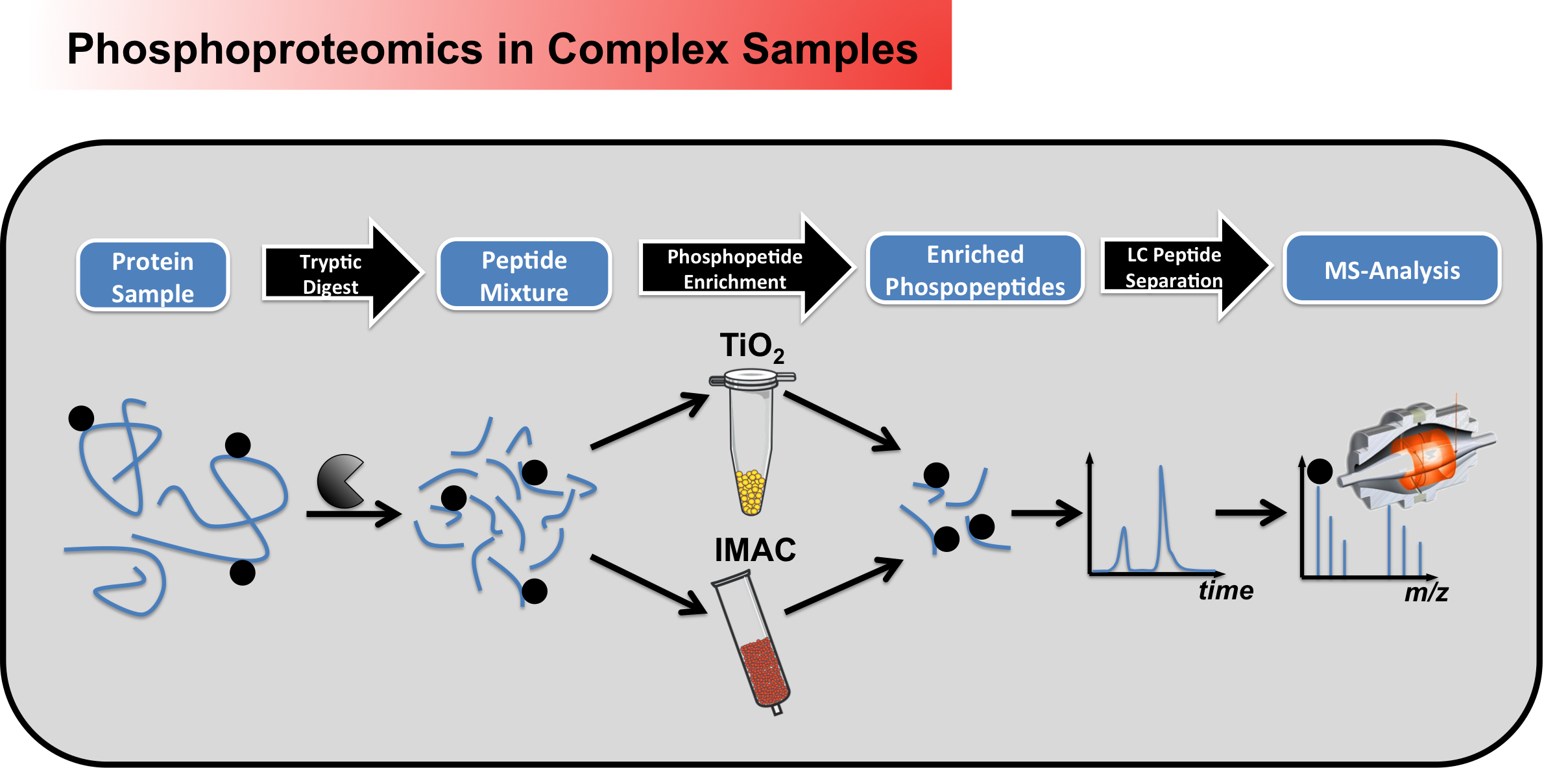 Figure 1 - Workflow for large scale phosphoproteomic screening in complex samples Beside discovery
driven determinations of
phosphorylation sites in a global
manner, specific phosphosites can
be monitored very sensitively
using parent mass list-based
approaches (Figure 2). Here the
masses of phosphopeptides of
interest are first calculated
in-silico, to allow a subsequent
targeted analysis within the mass
spectrometer.
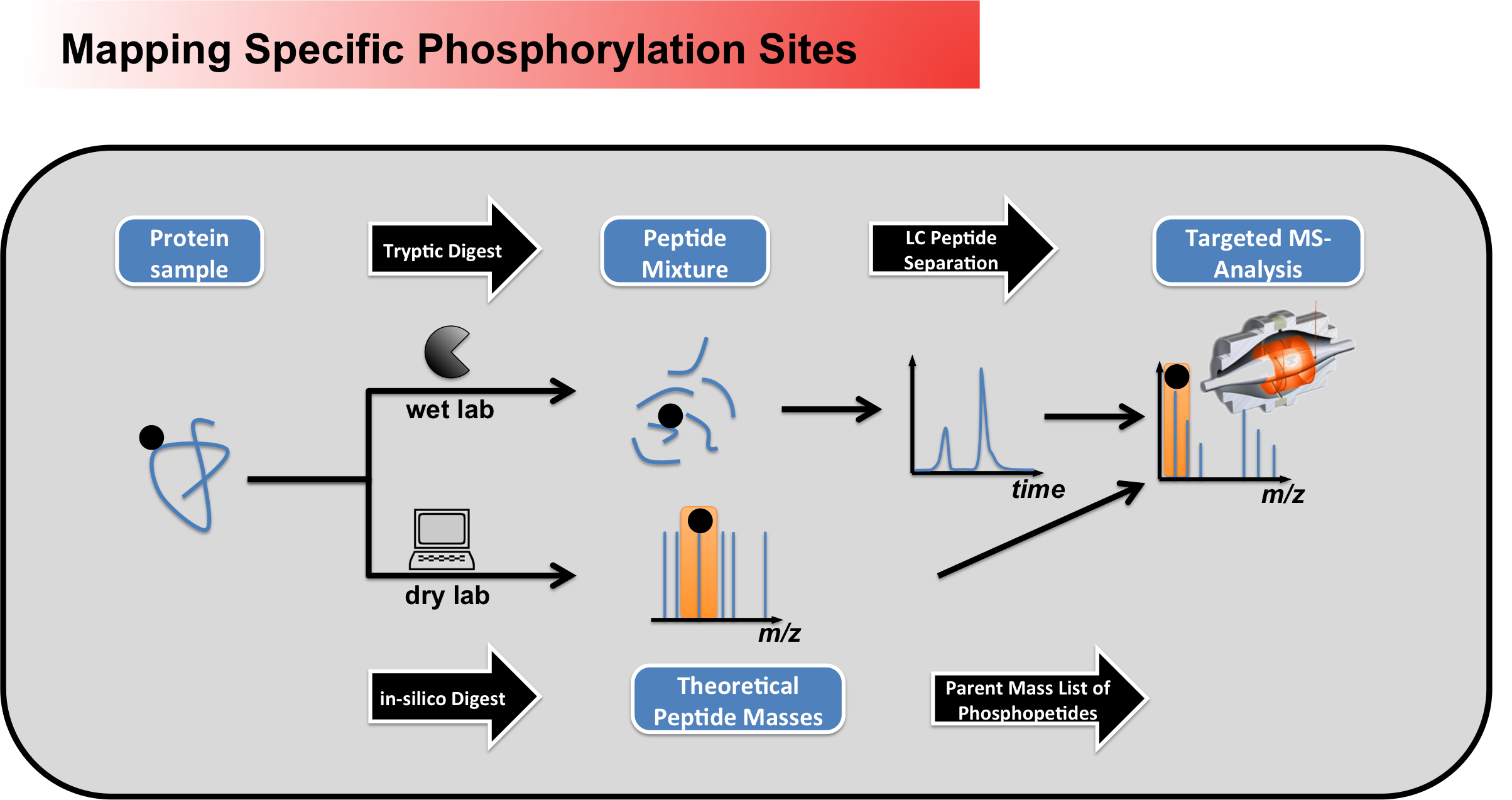 Figure 2 - Workflow for mapping specific phosphorylation sites using peptide parent mass lists Glykosylations We also perform
glykosylation analyses following
various strategies. For example,
together with the Strahl group we
established a protocol, which
combined peptide glykosylation,
lectin enrichment (ConA) and
differential mannosidase
treatment, to detect protein
O-mannosylation in a large scale
manner (see Figure 3 and
Winterhalter et al2).
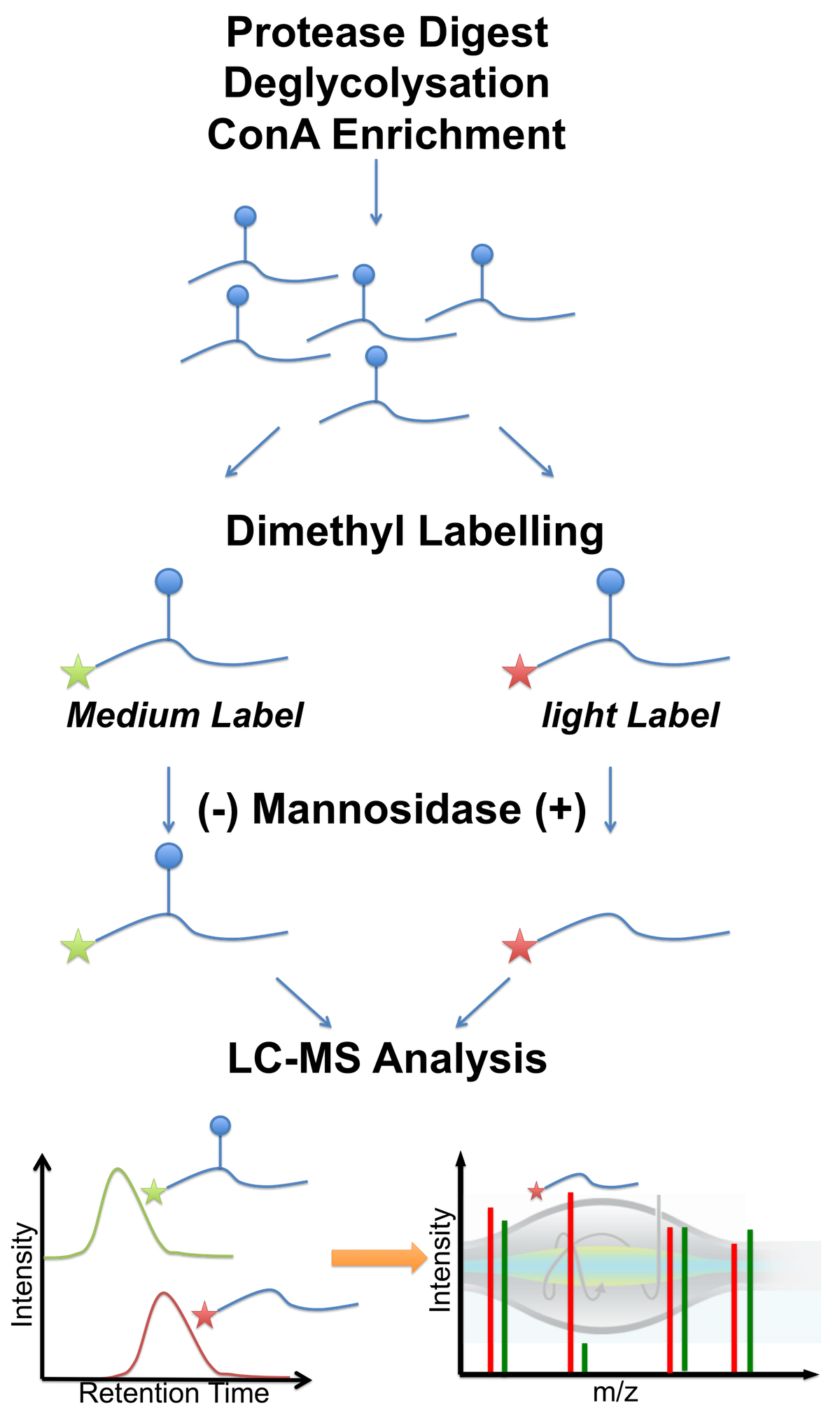 Figure 3 - Workflow of differential O-mannosylation analysis, including peptide deglycosylation, selective mannose peptide enrichment (lectin ConA), dimethyl labeling and differential mannosidase treatment. O-mannosylated peptides are identified by a characteristic shift in retention time, caused by the presence of the mannose group. Cystein redox states The analysis of
cystein redox states is also in
our portfolio. For this we apply
differential cystein labeling
techniques using common
cystein-reactive reagents, like
N-ethylmaleimide (NEM) or
iodoacetamide (IAA). Differential
labeling can either be performed with
isotopologues of the same
alkylation reagent (e.g. light vs
heavy NEM --> see Figure 4),
but alternatively also with two
different reagents (1. NEM
alkylation of free cysteines. 2.
Reduction of oxidized cysteines 3.
IAA alkylation of released
cysteines). Finally, this then
allows to determine
differences in the oxidative state
of cysteine sites in target
proteins both sensitively and
quantitatively.
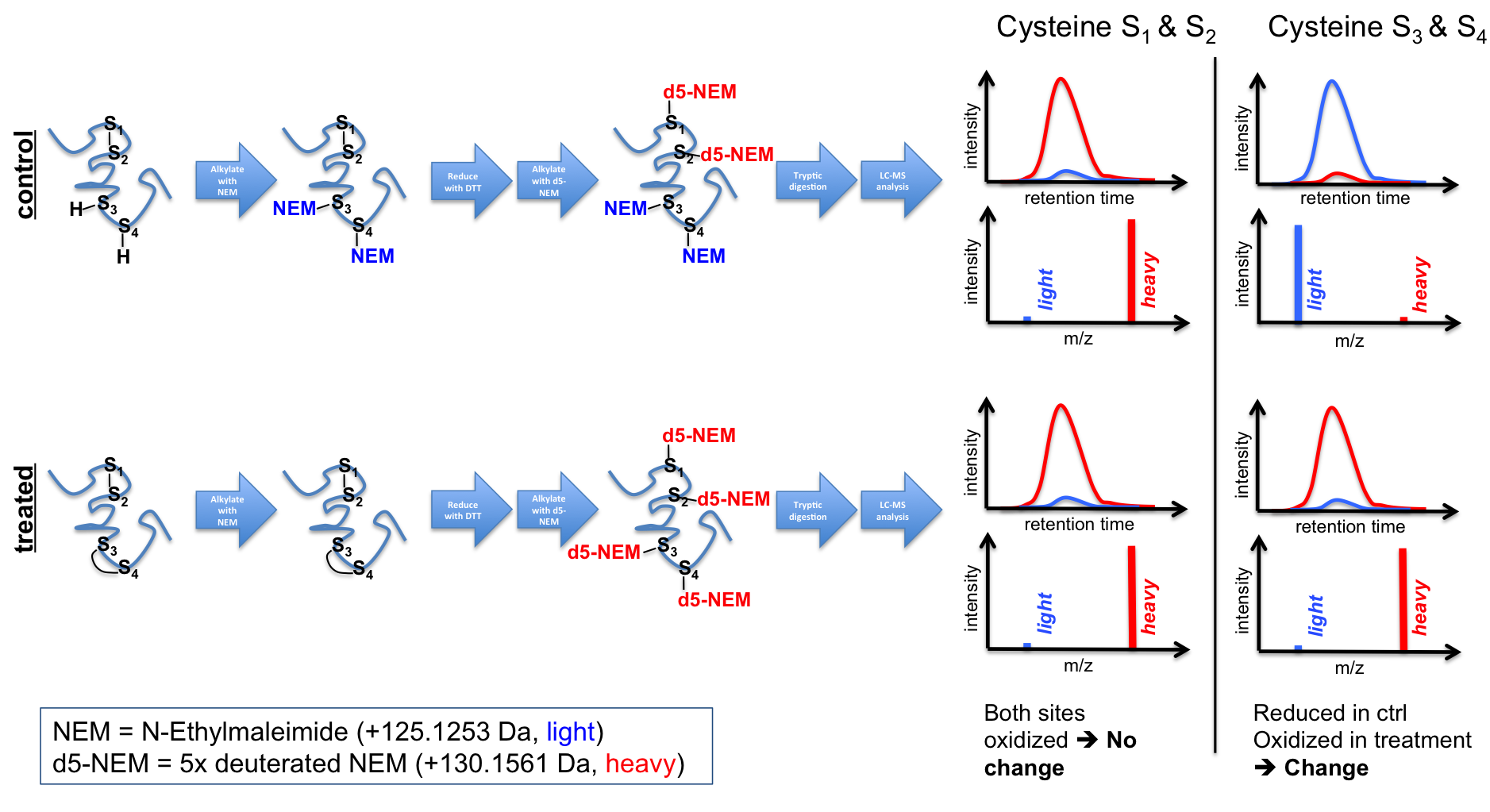 Figure 4 - Diagramm depicting the principal workflow of an isotope-based differential cysteine redox state analysis. References 1. Lin, T. C. et al. (2014) Cell-cycle dependent phosphorylation of yeast pericentrin regulates gamma-TuSC-mediated microtubule nucleation. Elife 3, e02208, doi:10.7554/eLife.02208 - Pubmed 2. Winterhalter, P. R., Lommel, M., Ruppert, T. & Strahl, S. (2013) O-glycosylation of the non-canonical T-cadherin from rabbit skeletal muscle by single mannose residues. FEBS letters 587, 3715-3721, doi:10.1016/j.febslet.2013.09.041 - Pubmed 3. Lommel, M. et al. (2013) Protein O-mannosylation is crucial for E-cadherin-mediated cell adhesion. Proc Natl Acad Sci U S A 110, 21024-21029, doi:10.1073/pnas.1316753110 4. Sobotta, M. C. et al. (2015) Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat Chem Biol 11, 64-70, doi:10.1038/nchembio.1695 - Pubmed |
|
||